Bank properties
A bank for BioMAJ mainly consists in several properties defined in a .properties file. The most useful properties will be presented here with a concrete example to clear things up.
Example of a <bank>.properties file:
Each bank owns its configuration file. It must be named bank_name.properties and located in conf.dir. The bank configuration file can override any global configuration expect above mandatory parameters.
The update process takes into account the last modified date of the bank config file. If file is updated, the whole process is restarted.
![]() It is possible to define variables and reuse them in the property file, example:
It is possible to define variables and reuse them in the property file, example:
myvar=myvalue
mysupervar=%(myvar)s/additional_value
# properties can be called with the syntax %(property_name)sOr to use variables from the bank file with syntax %(<variable>)s :
PROC0.args=%(data.dir)s/%(db.name)s %(mail.from)s %(mail.admin)sLet’s say we want to download alu bank from here:
Contents
- Basics
- Where to download the files from
- Where to donwload the files to
- Filtering files
- Meta information
- Additional mandatory properties
- Advanced notions
- Custom release number
- Reporting
- Properties organization
- More specific information and examples
Basics
We need to tell BioMAJ :
Where to download the files from
This is done thanks to 5 properties :
- protocol: tells what protocol to use to download the file. The protocol must not appear in the server property. In our case it is ftp. (example: ftp, http, local, multi)
- server: ftp, http, local, multi. In this example: ftp.ncbi.nlm.nih.gov
- remote.dir: path to the remote directory that contains the required files, note that the ‘/’ at the beginning is mandatory: /blast/db/FASTA/
- remote.files: contains one or more regular expressions that describe the files to retrieve. Let’s say we want the following files:
-
- All compressed alu files: ^alu.*\.gz$
- All README files: ^README.*
-
- server.credentials: Optional, credentials access to server, format= user:password
Final value is: ^alu.*\.gz$ ^README.*
Where to download the files to
3 properties are involved with this:
- dir.version: Directory, relative to data.dir where bank release will be stored, must not start with a / (in this example: ncbi/blast/alu)
- offline.dir.name: Directory to use for download, must be relative to data.dir and must not start with a / (offline/ncbi/blast/alu_tmp)
An additional property related to your repository management is keep.old.version. It basically tells how many versions of your bank you want BioMAJ to keep. If value is 1, BioMAJ will keep the current version and the previous one.
Which files to filter
- local.files: Once files have been downloaded and extracted (if needed) in data.dir/offline.dir.name directory, they are moved to the production
directory (data.dir/dir.version). You can tell BioMAJ to move only some of these files with that property. As for remote.files property, the value is a regular expression. It is possible to move all files : .* or files with a specific name, for example ending with a or n ^alu\.(a|n).*

Remote.file: Choose the directory structure of the downloaded files
- remote. file=genomes/fasta/.*\. gz: files saved in the offline directory by keeping the structure: offlinedir/genomes/fasta/
- remote. file=genomes/fasta/ (. *\. gz):files saved directly to: offlinedir/
- remote. file=genomes/ (fasta)/ (. *\. gz): files saved in: offlinedir/fasta
- remote. files = genomes/ (?: fasta|flat)/.*\. gz: regular expression management for the selection of offlinedir/genomes/fasta/ or offlinedir/genomes/flat/ files according to their origin
Remote.list : Create a list of files to download
It is possible to extract with a preprocess a list of files to download, using defined protocol, server etc. This file must be generated in json format:
- remote.list=path_to_file.json
- From BioMAJ version up to 3.0.20
- Remote.file should be empty
- Structure of the file must be compliant with the following definition:
[{"name": "alu.n.gz", "root": "/blast/db/FASTA/"}, {"name": "alu.n.gz.md5", "root": "/blast/db/FASTA/"}]- More information can be defined:
[{"save_as": "alu.n.gz", "group": "ftp", "name": "alu.n.gz", "month": 11, "user": "anonymous", "year": 2003, "hash": "027577640cd7f182826abbf83e76050c", "permissions": "-r--r--r--", "root": "/blast/db/FASTA/", "day": 26, "size": 24465}, {"save_as": "alu.n.gz.md5", "group": "ftp", "name": "alu.n.gz.md5", "month": 6, "user": "anonymous", "year": 2009, "hash": "98caf48b2f98cdf0c38bda8a74352266", "permissions": "-r--r--r--", "root": "/blast/db/FASTA/", "day": 15, "size": 43}]- Each additional field is optional.
If year AND month AND day are not defined, then release parameters must be defined in the properties to extract a release information. If not present, they will be set to current date, so a new release will be detected each day. If file is deleted by a post-process, then, at next run, file will not be detected and BioMAJ will consider there is no new release.
Meta information
Some properties that describe your bank :
- db.name: this property holds the name of the bank. uniprot in our case
- db.fullname: the field usually contains a description about the bank, for example: Some description…
- db.type: you can specify the bank type (comma separated). In our case it’s nucleic,protein.
Additional mandatory properties
So far, our file looks like:
db.name=alu
db.fullname="alu.n : alu repeat element. alu.a : translation of alu.n repeats"
db.type=nucleic,protein
server=ftp.ncbi.nlm.nih.gov
protocol=ftp
remote.dir=/blast/db/FASTA/
remote.files=^alu.*\.gz$ ^README.*$
local.files=.*
frequency.update=0
data.dir=/db/
offline.dir.name=offline/ncbi/blast/alu_tmp
dir.version=ncbi/blast/alu
keep.old.version=1Advanced notions
Custom release number
As we have not defined anything specific on how BioMAJ has to get the bank version, the default behaviour is to set as the release number the most recent file date on the server. In our case, that would be 26/11/2003 (format as defined in release.dateformat).
There are two other ways to do differently :
- release.file: Extraction of the release from the file name in this form
^filename_release_is_ ({d+_)_^. txt$ (For this to work, the definition of regexp for release.file needs to start with ‘^’ and end with ‘$’) - release.regexp: extraction of the release in the downloaded file (in case of the file in case the release number is not included in the file name). It is possible to use this version number to define a remote directory name in the following way: %(remoterelease)s/
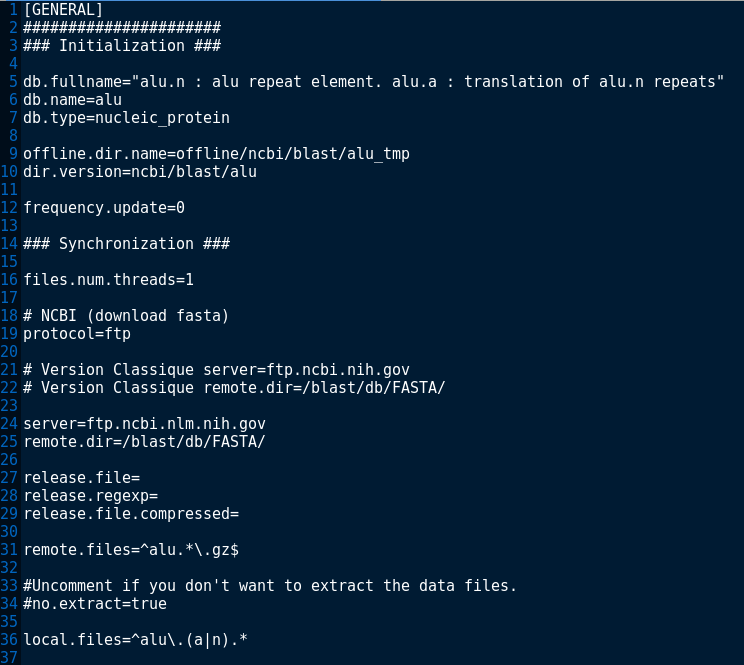
The file now looks like:
db.fullname="alu.n : alu repeat element. alu.a : translation of alu.n repeats"
db.name=alu
db.type=nucleic_protein
offline.dir.name=offline/ncbi/blast/alu_tmp
dir.version=ncbi/blast/alu
frequency.update=0
files.num.threads=1
protocol=ftp
server=ftp.ncbi.nlm.nih.gov
remote.dir=/blast/db/FASTA/
release.file=
release.regexp=
release.file.compressed=
remote.files=^alu.*\.gz$
#Uncomment if you don't want to extract the data files.
#no.extract=true
local.files=^alu\.(a|n).*It is possible to extract the release from a file located on another server than the server of the bank, with its own download parameters (if not specified BioMAJ will use the bank parameters protocol, server, remote.dir):
release.protocol=
release.server=
release.remote.dir=Computed banks
It is possible to create interdependent banks, for example A depends on B and C, B depends on D:
- The workflow of the main bank will check each dependency. If one of them needs updating, it will be updated first and then it will be the main bank’s turn.
- The main bank knows the location of the banks on which it depends and their release information.
- All banks are managed sequentially (no parallelization).
How to make banks depend on each other? (in the file of the main bank A.properties)
- depends=bankB: Bank A will receive a environment variable with path path to the bank B
- bankB.files.move=flat/*: copies all files to a subdirectory of A with the name of the bank B
Example:
#Main bank A.properties
dir.version=bankA/computed
depends=bankB
sub1.files.move=flat/*
In the file tree structure:
$ls bankA/computed/current/
flat bankB
$ls bankA/computed/current/bankB/flat
fileB_1.txt fileB_2.txt
How to do a direct download?
Direct URL definition: ftp or http
Example: http://test.org/test/test.fa
- protocol=directhttp
- server=test.org
- url.method=GET
- target.name=mytest.fa (save results as mytest.fa)
Example: http://test.org/test/test.fa?key1=val1,key2=val2
- protocol=directhttp
- server=test.org
- remote.dir=test/test.fa
- url.method=GET
- url.params=key1,key2
- key1.value=val1
- key2.value=val2
- target.name=mytest.fa
Example: ftp://test.org/test/test.fa
-
- protocol=directftp
- server=test.org
- remote.dir=test/test.fa
Reporting
- Logging level: You can change the logging level with the property historic.logfile.level with the following values : ERR, WARN, INFO, VERBOSE, DEBUG
- Mailing: BioMAJ can mail a report after each bank update. The subject of the mail contains 4 items:
- The bank name
- The workflow status STATUS[TRUE|FALSE] : TRUE means that everything went well (at least on BioMAJ point view), FALSE means that the process failed.
- The update status UPDATE[TRUE|FALSE] : TRUE means that a new version was found on the remote server. Note that it does not mean that the update succeeded, just that BioMAJ downloaded/will have to download new files. FALSE means that we already have the latest version.
- If UPDATE is TRUE, an item information is the version found on the remote server.
- To activate mail reporting, you have to fill in the following properties:
- mail.from: mail address of the sender
- mail.smtp.host: smtp server address
- mail.admin: list of mail addresses separated by commas the reports will be sent to.
How to check the log file of a databank?
You can consult the logs of a bank by reading the file:
cat <your path to biomaj data>/log/<bank>/<XXXXXXX>/bank.logThe number XXXXXXXXX corresponds to the download session number.
Example for the bank alu from:
[GENERAL]
######################
### Initialization ###
db.fullname="alu.n : alu repeat element. alu.a : translation of alu.n repeats"
db.name=alu_ori
db.type=nucleic_protein
offline.dir.name=offline/ncbi/alu_tmp
dir.version=ncbi/alu
frequency.update=0
### Synchronization ###
files.num.threads=1
# NCBI (download fasta)
protocol=ftp
server=ftp.ncbi.nih.gov
remote.dir=/blast/db/FASTA/
release.file=
release.regexp=
release.file.compressed=
remote.files=^alu.*\.gz$
#Uncomment if you don't want to extract the data files.
#no.extract=true
local.files=^alu\.(a|n).*
## Post Process ## The files should be located in the projectfiles/process directory
BLOCKS=BLOCK1
BLOCK1.db.post.process=META0
META0=test_biomaj
test_biomaj.name=biomaj_test
test_biomaj.desc=test biomaj
test_biomaj.cluster=false
test_biomaj.type=test
test_biomaj.exe=echo
test_biomaj.args="$datadir"
### Deployment ###
keep.old.version=1
More information
More example here.
Github documentation with more specific example here.
